Teamwork makes the dream work: the relationship that gave birth to us
- Miguel Gomez-Raya Vilanova

- Jun 21, 2024
- 7 min read
Updated: Feb 24, 2025
Some of the big questions of humankind are: Where do we come from? How did we come to be? Why did we come to be? Biology, in some way, tries to answer the same questions, how did life originate? Why did it originate? And while it is a difficult task going all the way back to the beginning, understanding why life evolved the way it did and how this was possible sheds some light on these big questions. Come with me on a little journey on one of the most important events in the history of evolution, an event that set up one of the biggest explosions in biodiversity Earth has ever seen. The birth of eukaryotes.

All living organisms are made of cells, some of us are pluricellular -composed of many cells-, like animals and plants, and others are unicellular -made of a single cell- like the bacterium Escherichia coli (1). However, our individual cells are very different from those of many microorganisms. On the one hand, our cells and those of some microorganisms are very complex, they have different compartments and contain a nucleus that encloses the genetic material and organelles like mitochondria -the powerhouse of our cells- or, in the case of plants and algae, plastids -which allow them to obtain energy from light. On the other hand, most microorganisms are much smaller and have no nucleus. This difference was one that biologists saw and used it for the early classifications of life1, (i) prokaryotes for those non-nucleated organisms (from ancient Greek “pró” -before- and “káruon” -nut) and (ii) eukaryotes for those with nucleus (“eu” -true). However, with time, new advancements in molecular biology started to arise and with them, new ideas were born. It was in the 70s when Carl R. Woese studied the ribosomal RNA of different microorganisms and realised that not all prokaryotic organisms were the same. Their cellular architecture looked very similar at a fast glance, but a closer look into their genes showed that indeed they were extremely different (2). They could be classified into two big groups. In this way, despite all arising from a single Last Universal Common Ancestor (LUCA), all living organisms can now be classified in three domains, (i) Bacteria and (ii) Archaea, which are prokaryotic, and (iii) Eucarya.
In the history of the evolution of life, however, these different types of organisms did not emerge at the same time. The first to split were Bacteria and Archaea, which in fact share many features like the basic cell architecture or the fact that their genomes are generally organised in a single circular chromosome, except for a few exceptions. Yet they still have many divergent traits, like the composition of their membranes or how their genomes are replicated or the information that they contain is processed3. Nevertheless, the more complex eukaryotic cell appeared much later in the evolutionary history of life in a process named eukaryogenesis, when the Last Eukaryotic Common Ancestor (LECA) was born1. How the LECA acquired such complexity and how such an important event for the diversity of life happened is still a hot topic for many biologists.
The first models on eukaryogenesis were popularised by Lynn Margulis (née Lynn Sagan), even though they initially were theorised much earlier (1). She hypothesised that both mitochondria -which now we know was present in the LECA- and plastids -found in those eukaryotes with photosynthetic capabilities- had a symbiotic origin. This indicates that their origin lay in a mutualistic relationship between two cells, a host and the future mitochondrion or plastid. The latter became more and more dependent on the former over time and lost their complexity finally to become the organelles that we find now. She developed this idea on the fact that, indeed, mitochondria and plastids have their own DNA and perform some independent functions from the cell, among other characteristics. The question was: which cells gave rise to those relationships? Molecular phylogenetic analyses of some proteins first helped identifying the probable ancestors of mitochondria and plastids (1). The answer being an alpha-proteobacterium and a cyanobacterium, respectively. With the later discovery of Archaea some scientists started to wonder if in fact the host of those bacteria was an archaeon (1),(3).
Current eukaryogenesis models should take into account all the molecular and phylogenetic knowledge acquired during many years of research. Recently, with the development of high-throughput sequencing techniques we have been able to generate a huge quantity of metagenomes, which are genomes of microorganisms sequenced directly from environmental samples (1). Metagenomics provides information from uncultured organisms, allowing to skip the most difficult and many times limiting step, the isolation of the microorganisms. Molecular tools, including metagenomics, have uncovered many new lineages and largely expanded our understanding of the microbial world, and in this expansion another surprising archaeal lineage was to be discovered, Asgardarchaeota (4). Asgardarchaeota is an archaeal phylum initially discovered in deep-sea sediments. The first genomes for members of this group were obtained in sediments from the vicinity of Loki’s castle hydrothermal vent, hence the name of the first Asgard archaeal group (Lokiarchaea). However, what is most interesting about this group of organisms is the fact that their genome encodes proteins that were thought to be specific to eukaryotes -known as Eukaryotic Signature Proteins (4). This characteristic, along with some phylogenetic analysis, supports the idea that the probable host during eukaryogenesis was indeed an Asgard archaeon. With all this information, current models follow this idea, but differ in the mechanisms and the reasons for this process (Figure 1) (1) In this short article we will focus on two of those theories: the inside-out and the syntrophy models.
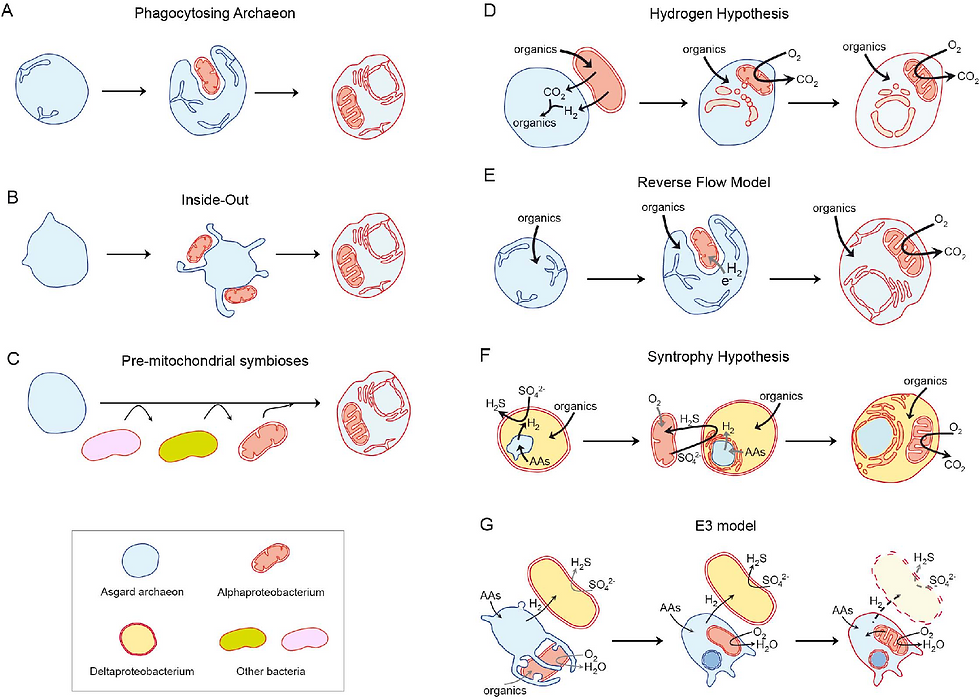
Figure 1
If you were to make an eukaryote following the inside-out recipe (5) you would just need an archaeal host with the capacity of creating protrusions in its membrane and a group of alpha-proteobacteria around it. These protrusions would, with time, surround the bacteria and engulf it until it became an integral part of the contents of the cell. This process would also form all the internal membranes and compartments eukaryotes have, as well as the nucleus. The isolation and culture of an Asgard archaeon and the imaging showing the presence of these protrusions supports the plausibility of this hypothesis (6). However, the membranes of archaea are very different in composition from those of eukaryotes, which are much similar to those of bacteria. Therefore, how did the membrane switch in composition? And why? are questions this theory cannot yet answer.
On the other hand, if you were to follow the syntrophy recipe (7) you would come into a very different scenario. For this recipe, you would need three ingredients: an Asgard archaeon, a sulphide-oxidising alpha-proteobacterium and a sulphate-reducing delta-proteobacterium. In this scenario, the relationship of the three microorganisms is maintained and needed because of their metabolic capabilities. The archaeon produces hydrogen that the delta-proteobacterium uses to reduce sulphate, the resulting sulphide is used by the alpha-proteobacterium who uses oxygen to oxidise it into sulphate again, closing the cycle. In this more detailed and complex model, the delta-proteobacterium would internalise the archaeon, who would become the nucleus of the cell, and then would acquire and internalise the alpha-proteobacterium. The isolation of another Asgard archaeon who indeed happens to bear a symbiotic relationship with a delta-proteobacterium, supports this hypothesis (8). Moreover, in this scenario the membrane composition would not need to be switched, as the external membrane would be that of a delta-proteobacterium, much more similar to the one of eukaryotes. In fact, a study of the virome of eukaryotes and an inference of the virome of the LECA would tend to support this scenario (9). However, if there were two endosymbiotic events why haven't we found yet any “eukaryotic” cells without mitochondria or mitochondria derived organelles? Moreover, we indeed find genes in eukaryotic genomes of delta-proteobacterial origin, but also from other bacterial phyla, were there other partners involved?
In summary, the complex process of eukaryogenesis is far from being uncovered. New discoveries are rapidly painting a clearer picture of what may have happened a couple of billion years ago. So hopefully soon we will be able to give a plausible answer to the how and why and shed some light into this mystery that has kept biologists wondering for so long.
References
1. López-García, P., and Moreira, D. (2023). The symbiotic origin of the eukaryotic cell. C R Biol. https://doi.org/10.5802/crbiol.118.
2. Woese, C.R., and Fox, G.E. (1977). Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.74.11.5088.
3. Lindås, A.-C., and Bernander, R. (2013). The cell cycle of archaea. Nat Rev Microbiol. https://doi.org/10.1038/nrmicro3077.
4. Spang, A., Saw, J.H., Jørgensen, S.L., Zaremba-Niedzwiedzka, K., Martijn, J., Lind, A.E., van Eijk, R., Schleper, C., Guy, L., and Ettema, T.J.G. (2015). Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. https://doi.org/10.1038/nature14447.
5. Baum, D.A., and Baum, B. (2014). An inside-out origin for the eukaryotic cell. BMC Biol. https://doi.org/10.1186/s12915-014-0076-2.
6. Rodrigues-Oliveira, T., Wollweber, F., Ponce-Toledo, R.I., Xu, J., Rittmann, S.K.-M.R., Klingl, A., Pilhofer, M., and Schleper, C. (2023). Actin cytoskeleton and complex cell architecture in an Asgard archaeon. Nature. https://doi.org/10.1038/s41586-022-05550-y.
7. López-García, P., and Moreira, D. (2020). The Syntrophy hypothesis for the origin of eukaryotes revisited. Nat Microbiol. https://doi.org/10.1038/s41564-020-0710-4.
8. Imachi, H., Nobu, M.K., Nakahara, N., Morono, Y., Ogawara, M., Takaki, Y., Takano, Y., Uematsu, K., Ikuta, T., Ito, M., et al. (2020). Isolation of an archaeon at the prokaryote-eukaryote interface. Nature. https://doi.org/10.1038/s41586-019-1916-6.
9. Krupovic, M., Dolja, V.V., and Koonin, E.V. (2023). The virome of the last eukaryotic common ancestor and eukaryogenesis. Nat Microbiol. https://doi.org/10.1038/s41564-023-01378-y
This article was copy edited by Kodie Noy.
Meet the author: Miguel Gomez-Raya Vilanova






Comments